Our Goal
Fed-BioMed is an open-source research and development initiative for translating collaborative learning into real-world medical applications, including federated learning and federated analytics.
The community of Fed-BioMed gathers experts in medical engineering, machine learning, communication, and security.
We all contribute to provide an open, user-friendly, and trusted framework for deploying the state-of-the-art of collaborative learning in sensitive environments, such as in hospitals and health data lakes.




Let's Start
Start using Fed-BioMed, follow our tutorials and user guide.
What's federated learning?
Discover the advantages of federated learning with Fed-BioMed
The goal of Federated learning is to allow collaborative learning with decentralized data.
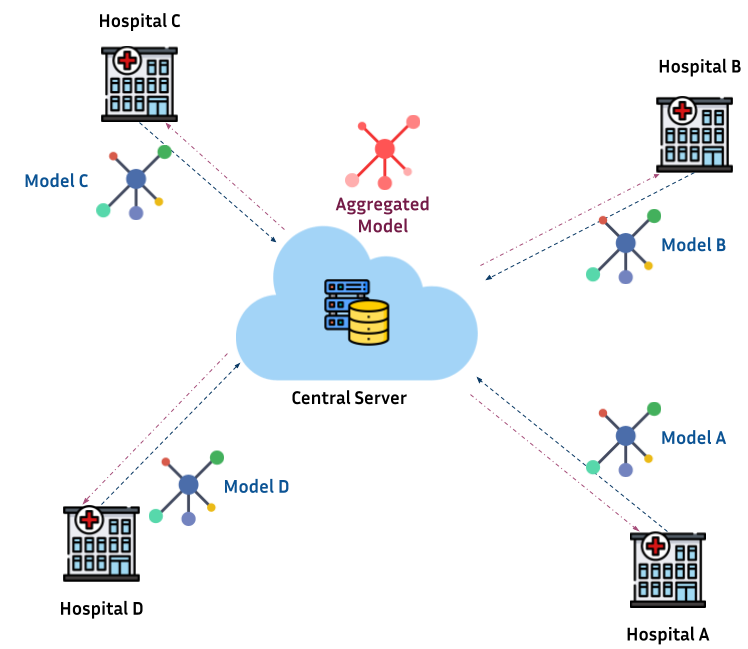
Healthcare is a typical application of federated learning: while hospitals across several geographical locations want to jointly train a machine learning model on the data hosted at each site, data cannot be shared between them because of privacy and security concerns. Federated learning gives us a methodological framework to train a global machine learning model, by only sharing the parameters of the models separately trained at each site. As a result, data never leaves the hospitals, and training is performed by simply aggregating models parameters to finally obtain a global model. Under certain conditions, the aggregated model faithfully represents the global variability across hospitals, and provides high generalization and robustness properties.
News

A new release of Fed-BioMed (v6.0) is now available!
A new major version of Fed-BioMed is now available, featuring a new API and more!
Read More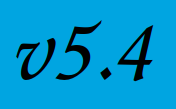
A new release of Fed-BioMed (v5.4) is out!
A new release of Fed-BioMed is now available, improving secure aggregation !
Read More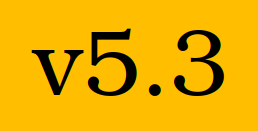
A new release of Fed-BioMed (v5.3) is out!
A new release of Fed-BioMed is now available, introducing LOM secure aggregation !
Read MoreFunding




Industrial Contributors
Users and Partners

Cite us!
If you use Fed-BioMed in your research, please cite our publication:
editor="Rehman, Muhammad Habib ur and Gaber, Mohamed Medhat",
title="Fed-BioMed: Open, Transparent and Trusted Federated Learning for Real-world Healthcare Applications",
bookTitle="Federated Learning Systems: Towards Privacy-Preserving Distributed AI",
year="2025",
publisher="Springer Nature Switzerland",
pages="19--41",
isbn="978-3-031-78841-3",
doi="10.1007/978-3-031-78841-3_2",
url="https://doi.org/10.1007/978-3-031-78841-3_2"
User support
Send a message to fedbiomed-support _at_ inria _dot_ fr
to benefit from the feedback of the community and/or
to request an invite to the Fed-BioMed support channel on Discord.
Please begin with checking the support list archive and issues archive for an existing answer to your problem.
When posting a support request, please pay attention to some tips:
- Be clear about what your problem is: what was the expected outcome,
what happened instead? Detail how someone else can recreate the problem. - Additional infos: link to demos, screenshots or code showing the problem.
You may also want to subscribe to the support list.
Contact Us
If you want to be part of Fed-BioMed contact fedbiomed _at_ inria _dot_ fr