Experiment Class of Fed-BioMed
Introduction
The Experiment class in Fed-BioMed is in charge of orchestrating the federated learning process on available nodes. Specifically, it takes care of:
- Searching the datasets on active nodes, based on specific tags given by a researcher and used by the nodes to identify the dataset.
- Sending model, training plan and training arguments to the nodes.
- Tracking the training process on the nodes during all training rounds.
- Checking the nodes' responses to handle possible failures.
- Receiving the local model parameters after every round of training.
- Aggregating the local model parameters based on the specified federated approach.
- Sending the aggregated parameters to the selected nodes for the next round.
- Optimizing the global model (i.e. the aggregated model).
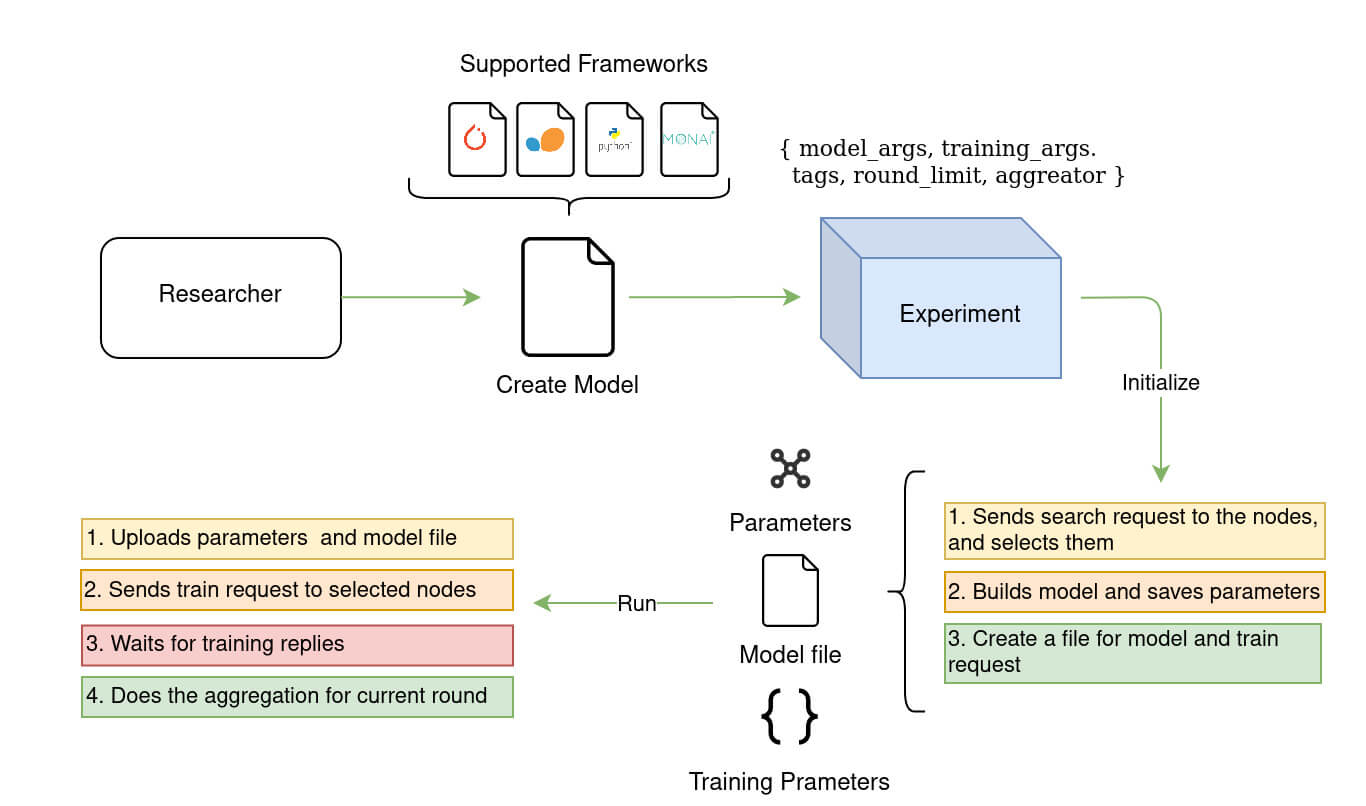
Defining an experiment
You may configure an Experiment by providing arguments to its constructor, as shown below.
from fedbiomed.researcher.federated_workflows import Experiment
exp = Experiment(tags=tags,
nodes=None,
training_plan_class=MyTrainingPlan,
training_args=training_args,
round_limit=rounds,
aggregator=FedAverage(),
node_selection_strategy=None)
It is also possible to define an empty experiment and set the arguments afterwards, using the setters of the experiment object. Please visit the tutorial In depth experiment configuration to find out more about declaring an experiment step by step
Under the hood, the Experiment class takes care of a lot of heavy lifting for you. For example, when you initialize an experiment with the tags argument, it uses them to automatically create a FederatedDataSet by querying the federation. Afterwards, Experiment initializes several internal variables to manage federated training on all participating nodes. Finally, it also creates the strategy to select the nodes for each training round. When the node_selection_strategy is set to None, the experiment uses the default strategy which is DefaultStrategy.
Setting the training data
Setting the training data by setting the tags
Each dataset deployed on the nodes is identified by tags. Tags allow researchers to select the same dataset registered under a given tag (or list of tags) on each node for the training.
The argument tags of the experiment is used for dataset search request. It can be a list of tags which are of type string, or single tag of type string.
exp = Experiment()
exp.set_tags(tags=['#MNIST', '#dataset'])
#or
exp.set_tags(tags='#MNIST')
Setting tags also sets the Experiment's training data
Whenever the set_tags method is called, a query is always issued to identify all nodes in the federation that have datasets with matching tags. Consequently, the training data of Experiment is changed to match the results from the query.
You can check your tags in your experiment as follows:
tags = exp.tags()
print(tags)
# > OUTPUT:
# > ['#MNIST', '#dataset']
Tags matching multiple datasets
An Experiment object must have one unique dataset per node. Object creation fails if this is not the case when trying to instantiate the FederatedDataSet object. This is done to ensure that training for an Experiment uses only a single dataset for each node.
As a consequence, tags specified for an Experiment should not be ambiguous, which means they cannot match multiple datasets on one node.
For example if you instantiate Experiment(tags='#dataset') and a node has registered one dataset with tags ['#dataset', '#MNIST'] and another dataset with tags ['#dataset', '#foo'] then experiment creation fails.
Setting the training data by providing the metadata directly
The dataset metadata can be provided directly using the set_training_data method. The metadata can be a FederatedDataSet object or a nested dict with format {node_id: {metadata_key: metadata_value}}.
When you provide a metadata object directly, the Experiment's tags attribute is set to None.
Under-the-hood consistency with all members of Experiment
When you change the training data (either through set_tags or set_training_data), the Experiment class performs a lot of operations to ensure that consistency is maintained for all of its attributes that use the training data. In particular, the aggregator and node_state_agent classes are updated with the new training data.
Selecting specific Nodes for the training
The argument nodes stands for filtering the nodes that are going to be used for federated training. It is useful when there are too many nodes on the network, and you want to perform federated training on specific ones. nodes argument is a list that contains node ids. When it is set, the experiment queries only the nodes that are in the list for a dataset matching tags. You can visit listing datasets and selecting nodes documentation to get more information about this feature.
nodes = ['node-id-1', 'node-id-2']
exp.set_nodes(nodes=nodes)
By default, nodes argument is None which means that each node that has a registered dataset matching all the tags will be part of the federated training.
exp.set_nodes(nodes=None)
Node filtering happens at training time
Setting nodes doesn't mean sending another dataset search request to the nodes. Node filtering happens dynamically each time a training request is sent to nodes. In other words, if you search again for datasets after setting nodes by running exp.set_training_data(training_data=None, from_tags=True) you select in your FederatedDataset the same nodes as with nodes=None.
Load your Training Plan: Training Plan Class
The training_plan_class is the class where the model, training data and training step are defined. Although not required, optimizers and dependencies can also be defined in this class. The experiment will extract source of your training plan, save as a python module (script), and send the source code to the nodes every round of training. Thanks to that, each node can construct the model and perform the training.
class MyTrainingPlan(TorchTrainingPlan):
def init_model(self, model_args):
# Builds the model and returns it
return model
def init_optimizer(self, optimizer_args):
# Builds the optimizer and returns it
return optimizer
def training_step(self):
# Training step at each iteration of training
return loss
def training_data(self):
# Loads the dat and returns Fed-BioMed DataManager
return DataManager()
exp.set_training_plan_class(training_plan_class=MyTrainingPlan)
# Retrieving training plan class from experiment object
training_plan_class = exp.training_plan_class()
Model Arguments
The model_args is a dictionary with the arguments related to the model (e.g. number of layers, layer arguments and dimensions, etc.). This will be passed to the init_model method during model setup. An example for passing the number of input and output features for a model is shown below.
model_args = {
"in_features" : 15,
"out_features" : 1
}
exp.set_model_args(model_args=model_args)
Incompatible model_args
If you try to set new model_args that are incompatible with the current model weights, the function will raise an exception and the Experiment class will be left in an inconsistent state. To rectify this, immediately re-execute set_model_args with additional keyword argument keep_weights=False as in the example below:
exp.set_model_args(model_args, keep_weights=False)
Model arguments can then be used within a TrainingPlan as in the example below,
class MyTrainingPlan(TorchTrainingPlan):
def init_model(self, model_args):
model = self.Net(model_args)
return model
class Net(nn.Module):
def __init__(self, model_args):
super().__init__()
self.in_features = model_args['in_features']
self.out_features = model_args['out_features']
self.fc1 = nn.Linear(self.in_features, 5)
self.fc2 = nn.Linear(5, self.out_features)
Special model arguments for scikit-learn experiments
In scikit-learn experiments, you are required to provide additional special arguments in the model_args dictionary. For classification tasks, you must provide both a n_features and an n_classes field, while for regression tasks you are only required to provide a n_features field.
n_features: an integer indicating the number of input features for your modeln_classes: an integer indicating the number of classes in your task
Note that, as an additional requirement for classification tasks, classes must be identified by integers in the range 0..n_classes
Training Arguments
training_args is a dictionary, containing the arguments for the training on the node side (e.g. data loader arguments, optimizer arguments, epochs, etc.). These arguments are dynamic, in the sense that you may change them between two rounds of the same experiment, and the updated changes will be taken into account (provided you also update the experiment using the set_training_args method).
A list of valid arguments is given in the TrainingArgs.default_scheme documentation.
To set the training arguments you may either pass them to the Experiment constructor, or set them on an instance with the set_training_arguments method:
exp.set_training_arguments(training_args=training_args)
exp.training_args()
Controlling the number of training loop iterations
The preferred way is to set the num_updates training argument. This argument is equal to the number of iterations to be performed in the training loop. In mini-batch based scenarios, this corresponds to the number of updates to the model parameters, hence the name. In PyTorch notation, this is equivalent to the number of calls to optimizer.step.
Another way to determine the number of training loop iterations is to set epochs in the training arguments. In this case, you may optionally set a batch_maxnum argument, in order to exit the training loop before a full epoch is completed. If batch_maxnum is set and is greater than 0, then only batch_maxnum iterations will be performed per epoch.
num_updates is the same for all nodes
If you specify num_updates in your training_args, then every node will perform the same number of updates. Conversely, if you specify epochs, then each node may perform a different number of iterations if their local dataset sizes differ.
Note that if you set both num_updates and epochs by mistake, the value of num_updates takes precedence, and epochs will effectively be ignored.
Why num updates?
In a federated scenario, different nodes may have datasets with very different sizes. By performing the same number of epochs on each node, we would be biasing the global model towards the larger nodes, because we would be performing more gradient updates based on their data. Instead, we fix the number of gradient updates regardless of the dataset size with the num_updates parameter.
Compatibility
Not all configurations are compatible with all types of aggregators and experiments. We list here the known constraints:
- the Scaffold aggregator requires using
num_updates
Batch size and other data loader arguments
Dataloader arguments are automatically injected through the DataManager class
It is strongly recommended to always provide a loader_args key in your training arguments, with a dictionary as value containing at least the batch_size key.
Example of minimal loader arguments:
training_args = {
'loader_args': {
'batch_size': 1,
},
}
Note that the loader_arguments, as well as any additional keyword arguments that you will specify in your DataManager constructor, will be automatically injected in the definition of the data loader. Please refer to the training_data method documentation for more details.
Setting a random seed for reproducibility
The random_seed argument allows to set a random seed at the beginning of each round.
random_seed is set both on the node and the researcher
The random_seed is set whenever the TrainingPlan is instantiated: both on the researcher side before sending the train command for a new round, and on the node side at the beginning of the configuration of the new training round.
Setting the random_seed affects:
- the random initialization of model parameters at the beginning of the experiment
- the random splitting of data between training and testing subset
- the random shuffling of data in the
DataLoader - any other random effect on the node and researcher side.
The same seed is used for the built-in random module, numpy.random and torch.random, effectively equivalent to:
import random
import numpy as np
import torch
random.seed(training_args['random_seed'])
np.random.seed(training_args['random_seed'])
torch.manual_seed(training_args['random_seed'])
random_seed is reset at every round
The random_seed argument will be reset to the specified value at the beginning of every round.
Because of this, the same sequence will be used for random effects like shuffling the dataset for all the rounds within an exp.run() execution. This is not what the user typically wants. A simple workaround is to manually change the seed at every round and use exp.run_once instead:
for i in range(num_rounds):
training_args['random_seed'] = 42 + i
exp.set_training_args(training_args)
exp.run_once()
Sub-arguments for optimizer and differential privacy
In Pytorch experiments, you may include sub arguments such as optimizer_args and dp_args. Optimizer arguments represents the arguments that are going to be passed to def init_optimizer(self, o_args) method as dictionary and (dp_args) represents the arguments of differential privacy.
Optimizer arguments and differential privacy arguments are valid only in PyTorch base training plan.
training_args = {
'loader_args': {
'batch_size': 20,
},
'num_updates': 100,
'optimizer_args': {
'lr': 1e-3,
},
'dp_args': {
'type': 'local',
'sigma': 0.4,
'clip': 0.005
},
'dry_run': False,
}
Sharing persistent buffers
In Pytorch experiments, you may include the argument share_persistent_buffers. When set to True (default), nodes will share the full state_dict() of the Pytorch module, which contains both the learnable parameters and the persistent buffers (defined as invariant in the network, like batchnorm’s running_mean and running_var). When set to False, nodes will only share learnable parameters.
This argument will be ignored for scikit-learn experiments, as the notion of persistent buffers is specific to Pytorch.
Aggregator
An aggregator is one of the required arguments for the experiment. It is used on the researcher for aggregating model parameters that are received from the nodes after every round. By default, when the experiment is initialized without passing any aggregator, it will automatically use the default FedAverage aggregator class. However, it is also possible to set a different aggregation algorithm with the method set_aggregator. Currently, Fed-BioMed has only FedAverage and Scaffold classes, but it is possible to create a custom aggregator class. You can see the current aggregator by running exp.aggregator(). It will return the aggregator object that will be used for aggregation.
When you pass the aggregator argument as None it will use FedAverage aggregator (performing a Federated Averaging aggregation) by default.
exp.set_aggregator(aggregator=None)
or you can directly pass an aggregator instance
from fedbiomed.researcher.aggregators.fedavg import FedAverage
exp.set_aggregator(aggregator=FedAverage())
Custom aggregator classes should inherit from the base class Aggregator of Fed-BioMed. Please visit user guide for aggregators for more information.
About Scaffold Aggregator
FedAverage reflects only how local models sent back by Nodes are aggregated, whereas Scaffold also implement additional elements such as the Optimizer on Researcher side. Please note that currently only FedAverage is compatible with declearn's Optimizers.
Node Selection Strategy
Node selection Strategy is also one of the required arguments for the experiment. It is used for selecting nodes before each round of training. Since the strategy will be used for selecting nodes, thus, training data should be already set before setting any strategies. Then, strategy will be able to select among training nodes that are currently available regarding their dataset.
By default, set_strategy(node_selection_strategy=None) will use the default DefaultStrategy strategy. It is the default strategy in Fed-BioMed that selects for the training all the nodes available regardless their datasets. However, it is also possible to set different strategies. Currently, Fed-BioMed only provides DefaultStrategy but you can create your custom strategy classes.
Round Limit
The experiment should have a round limit that specifies the max number of training round. By default, it is None, and it needs to be created either declaring/building experiment class or using setter method for round limit. Setting round limit doesn't mean that it is going to be permanent. It can be changed after running the experiment once or more.
exp.set_round_limit(round_limit=4)
To see current round limit of the experiment:
exp.round_limit()
You might also wonder how many rounds have been completed in the experiment. The method round_current() will return the last round that has been completed.
exp.round_currrent()
Displaying training loss values through Tensorboard
The argument tensorboard is of type boolean, and it is used for activating tensorboard during the training. When it is True the loss values received from each node will be written into tensorboard event files in order to display training loss function on the tensorboard interface.
Tensorboard events are controlled by the class called Monitor. To enable tensorboard after the experiment has already been initialized, you can use the method set_monitor() of the experiment object.
exp.set_monitor(tensorboard=True)
You can visit tensorboard documentation page to get more information about how to use tensorboard with Fed-BioMed
Saving Breakpoints
Breakpoint is a researcher side function that saves an intermediate status and training results of an experiment to disk files. The argument save_breakpoints is of type boolean, and it indicates whether breakpoints of the experiment should be saved after each round of training or not. save_breakpoints can be declared while creating the experiment or after using its setter method.
exp.set_save_breakpoints(True)
Info
Setting save_breakpoints to True after the experiment has performed several rounds of training will only save the breakpoints for remaining rounds.
Please visit the tutorial "Breakpoints (experiment saving facility)" to find out more about breakpoints.
Experimentation Folder
Experimentation folder indicates the name of the folder in which all the experiment results will be stored/saved. By default, it will be Experiment_XXXX, and XXXX part stands for the auto increment (hence, first folder will be named Experiment_0001, the second one Experiment_0002 and so on). However, you can also define your custom experimentation folder name.
Passing experimentation folder while creating the experiment;
exp = Experiment(
#....
experimentation_folder='MyExperiment'
#...
)
Setting experimentation folder using setter;
exp.set_experimentation_folder(experimentation_folder='MyExperiment')
Using custom folder name for your experimentation might be useful for identifying different types of experiment. Experiment folders will be located at ${FEDBIOMED_DIR}/var/experiments. However, you can always get exact path to your experiment folder using the getter method experimentation_path(). Below is presented a way to retrieve all the files from the folder using os builtin package.
import os
exp_path = exp.experimentation_path()
os.listdir(exp_path)
Running an Experiment
train_request and train_reply messages
Running an experiment means starting the training process by sending train request to nodes. It creates training request that are subscribed by each live node that has the dataset. After sending training commands it waits for the responses that will be sent by the nodes. The following code snippet represents an example of train request.
{
"researcher_id": "researcher id that sends training command",
"experiment_id": "created experiment id by experiment",
"state_id": "state id for this round this experiment on this node",
"training_args": {
"loader_args": {
"batch_size": 32
},
"optimizer_args": {
"lr": 0.001
},
"epochs": 1,
"dry_run": false,
"batch_maxnum": 100
},
"dataset_id": "id of the used dataset on this node",
"training": True,
"model_args": <args>,
"params": <model weights>,
"training_plan": "<training plan code>",
"training_plan_class": "MyTrainingPlan",
"command": "train",
"round": <round_number>,
"aggregator_args": <args>,
"aux_vars": [list of auxiliary variables],
"secagg_arguments": {
"secagg_servkey_id": "secure aggregation server key id",
"secagg_random": <random number>,
"secagg_clipping_range": 3
]
}
}
}
After sending train requests, Experiment waits for the replies that are going to be published by each node once every round of training is completed. These replies are called training replies, and they include information about the training and the URL from which to download model parameters that have been uploaded by the nodes to the file repository. The following code snippet shows an example of training_reply from a node.
{
"researcher_id":"researcher id that sends the training command",
"experiment_id":"experiment id that creates training request",
"success":True,
"node_id":"ID of the node that completes the training ",
"dataset_id":"dataset_dcf88a68-7f66-4b60-9b65-db09c6d970ee",
"timing":{
"rtime_training":87.74385611899197,
"ptime_training":330.388954968
},
"msg":"",
"command":"train",
"state_id": "state id for new round this experiment on this node",
"params": <model weights>,
...
}
training_reply always results of a training_request sent by the Researcher to the Node. To complete one round of training, the experiment waits until receiving each reply from nodes. At the end of the round, it downloads the model parameters that are indicated in the training replies. It aggregates the model parameters based on a given aggregation class/algorithm. This process is repeated until every round is completed. Please see Figure 1 to understand how federated training is performed between the nodes and the Researcher (Experiment) component.
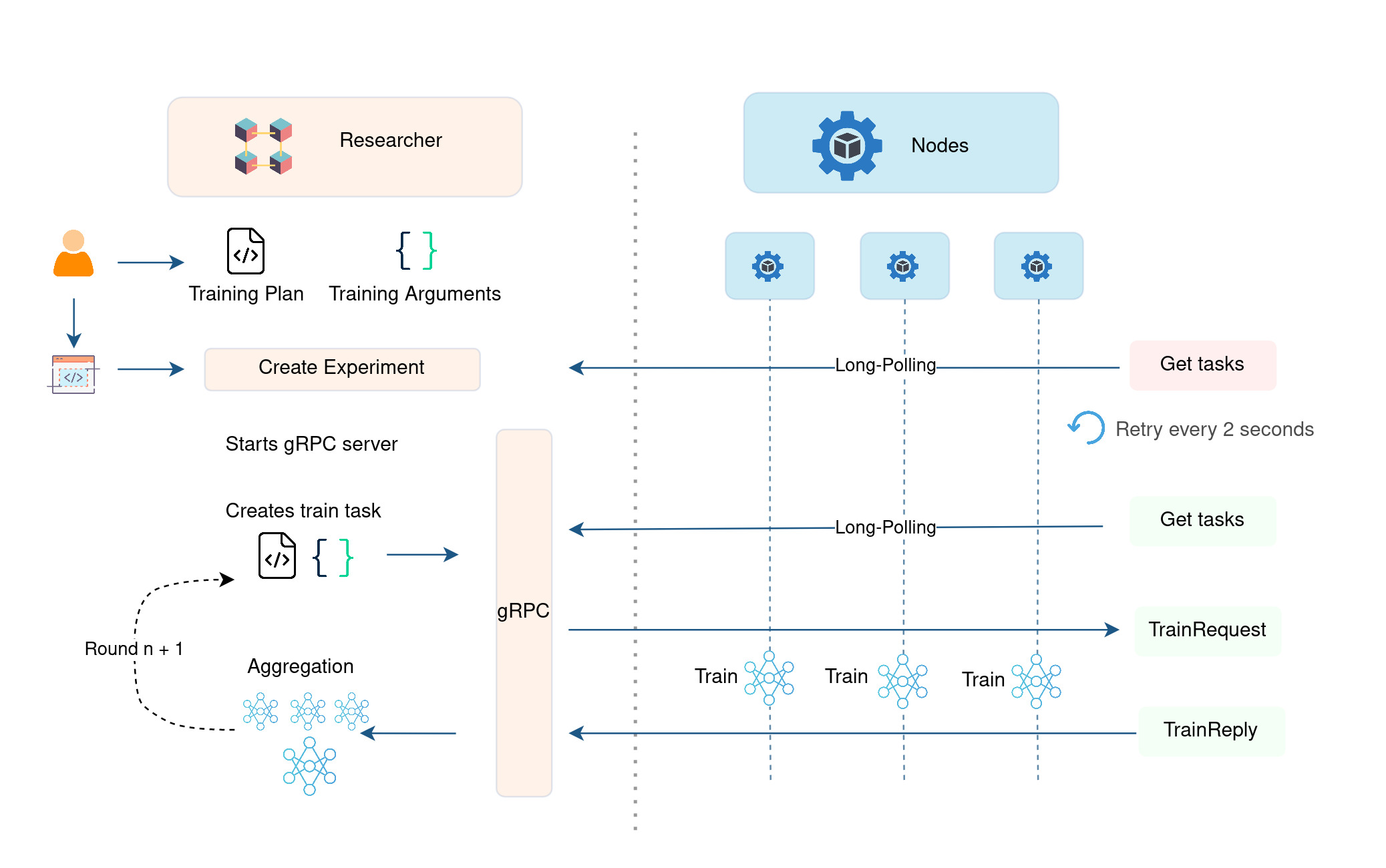 Figure 2 - Federated training workflow among the components of Fed-BioMed. It illustrates the messages exchanged between
Figure 2 - Federated training workflow among the components of Fed-BioMed. It illustrates the messages exchanged between Researcher and 2 Nodes during a Federated Training
The Methods run()and run_once()
In order to provide more control over the training rounds, Experiment class has two methods as run and run_once to run training rounds.
run()runs the experiment rounds from current round to round limit. If the round limit is reached it will indicate that the round limit has been reached. However, the methodruntakes 2 arguments asroundsandincrease.roundsis an integer that indicates number of rounds that are going to be run. If the experiment is at round0, the round limit is4, and if you passroundsas 3, it will run the experiment only for3rounds.increaseis a boolean that indicates whether round limit should be increased if the givenroundspass over the round limit. For example, if the current round is3, the round limit is4, and theroundsargument is2, the experiment will increase round limit to5
run_once()runs the experiment for single round of training. If the round limit is reached it will indicate that the round limit has been reached. This command is the same asrun(rounds=1, increase=False). However, ifrun_onceis executed asrun_once(increase=True), then, when the round limit is reached, it increases the round limit for one extra round.
To run your experiment until the round limit;
exp.run()
To run your experiment for given number of rounds (while not passing the round limit):
exp.run(rounds=2)
To run your experiment for given number of rounds and increase the round limit accordingly if needed:
exp.run(rounds=2, increase=True)
To run your experiment only once (while not passing the round limit):
exp.run_once()
To run your experiment only once even round limit is reached (and increase the round limit accordingly if needed):
exp.run_once(increase=True)
Running experiment with both run(rounds=rounds, increase=True) and run_once(increase=True) will automatically increase/update round limit if it is exceeded.


