Model Validation During Federated Training on the Nodes
Model validation is critical to discover how the model performs during the training rounds, when no dedicated holdout dataset is available for testing. In federated training, models are refined on the different nodes with different datasets. Therefore, model validation should be implemented on each node separately to compare model performances after its parameters are updated. Fed-BioMed provides a validation routine on the datasets that are randomly split at each round of training.
In the federated learning concept, two validation types can be applied at each round of training:
- Validation on globally updated parameters (
test_on_global_updates): It is the validation applied on the aggregated parameters before performing the training for the current round on a node. - Validation on locally updated parameters (
test_on_local_updates): It is the validation applied after the local training for the current round is performed on a node, and model parameters have been locally updated.
These two validations allow the users to compare how training on the node has improved the model performance.
Validation and training dataset are kept separated when testing is enabled
Newest Fed-BioMed releases (>=6.1) allows Researchers to keep distincts validation (for validating model performance) and training dataset when running the Experiment. It is now the default behaviour. In previous versions of Fed-BioMed, Nodes did not provide completely separated datasets for validating model performance. Since the samples for validation and training were picked randomly at each round of training, the same samples could be used for training in one round and for validation in another round. To revert back to this old behaviour, you can set in the Experiment.set_training_args shuffle_testing_dataset=True (defaults to False). Setting shuffle_testing_dataset=True could make sense when dataset are very small, and you may want to use the whole dataset to train your model, validation samples included.
Validation dataset are re-shuffled if test_ratio changes from one Round to another
Changing the value of test_ratio from one Round to another automatically re-shuffles the validation and testing dataset. It is equivalent to setting shuffle_testing_dataset=True.
Figure 1 illustrates the phases of validation and training during 2 rounds of federated training. As it can be seen in the figure, after the last round of training, one last validation on global updates is performed on the last aggregated parameters by each node. Therefore, the number of validation on globally updated parameters, if it is activated, will be equal to the number of rounds + 1
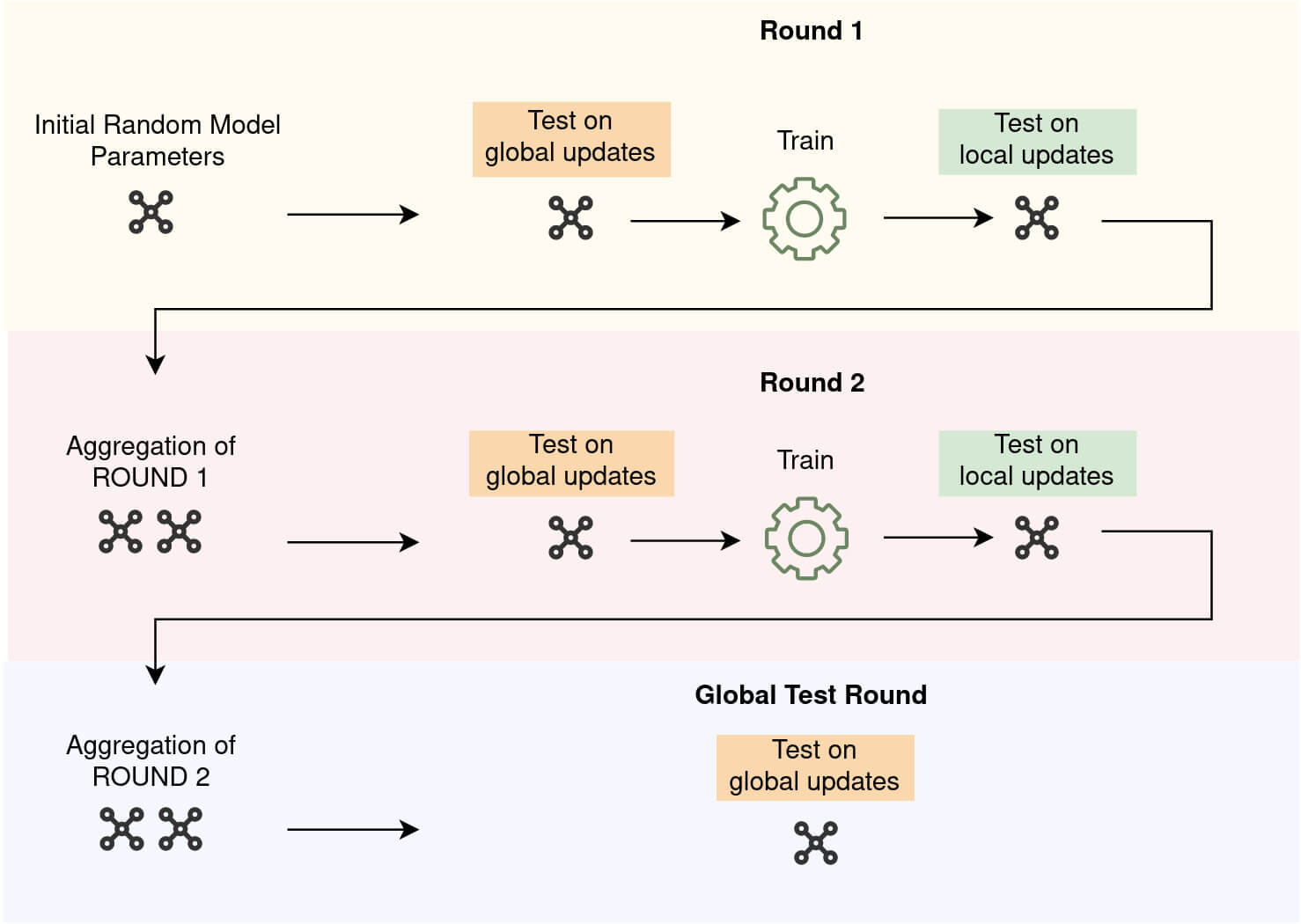 Figure 1 - Validation on global and local updates
Figure 1 - Validation on global and local updates
Default Metrics Provided by Fed-BioMed
Fed-BioMed provides several test metrics to perform validation that can be used without defining a validation function in the training plan. It allows the user to launch an experiment by providing as less code as possible. You can display all the metric provided by Fed-BioMed as shown in the following code snippet.
from fedbiomed.common.metrics import MetricTypes
MetricTypes.get_all_metrics()
# > Output:
# > ['ACCURACY', 'F1_SCORE', 'PRECISION', 'RECALL',
# > 'MEAN_SQUARE_ERROR', 'MEAN_ABSOLUTE_ERROR',
# > 'EXPLAINED_VARIANCE']
Note
By default, ACCURACY metric is used as a test metric if there isn't a metric defined by the researcher. Therefore, please pay attention to whether ACCURACY is relevant for the model that is going to be trained. Otherwise, metric results might be inconsistent.
Validation Arguments
Validation during the training is an optional process, and validation arguments should be configured in order to activate it. Here is the list of validation arguments that can be configured.
test_ratio: Ratio of the validation partition of the dataset. The remaining samples will be used for training. By default, it is0.0.test_on_global_updates: Boolean value that indicates whether validation will be applied to globally updated (aggregated) parameters (see Figure 1). Default isFalsetest_on_local_updates: Boolean value that indicates whether validation will be applied to locally updated (trained) parameters (see Figure 1). Default isFalsetest_metric: One ofMetricTypesthat indicates which metric will be used for validation. It can bestror an instance ofMetricTypes(e.g.MetricTypes.RECALLorRECALL). If it isNoneand there isn'ttesting_stepdefined in the training plan (see section: Define Custom Validation Step) default metric will beACCURACY.test_metric_args: A dictionary that contains the arguments that will be used for the metric function.test_batch_size: A value used to compute metrics using batches instead of loading the full testing dataset (specified bytest_ratio). Settingtest_batch_sizecan avoid havingMemoryErrorerrors due to large and /or heavy datasets. You should select wisely the batch size and the metric, for some metrics can be meaningless if computed over several small batches of data (e.g. explained variance).test_batch_sizeshould be greater or equal than1(enabled) or equal to0orNone(disabled).shuffle_testing_dataset: This argument will perform shuffling of the dataset. If it is switched fromFalsetooTrueit will reinitialize testing dataset that will be different than the one used in the previous rounds.
Info
Validation functions for each default metric executes functions from scikit-learn framework. Therefore, test_metric_args should be coherent with the arguments of "scikit-learn" metrics functions. Please visit here to see API documentation of scikit-learn metrics.
To activate validation on the node side, the arguments test_ratio and at least one of test_on_local_updates or test_on_global_updates should be set to True. Since the default values of test_on_local_updates and test_on_global_updates are False, setting test_ratio will only split dataset as validation and train sets but won't perform validation.
Setting Validation Arguments in Training Arguments
Validation arguments are considered a part of the training on the node side. Therefore, it is possible to define validation arguments in the training arguments and pass them to the experiment.
from fedbiomed.common.metrics import MetricTypes
from fedbiomed.researcher.federated_workflows import Experiment
training_args = {
#....
'optimizer_args': {
'lr': 1e-3
},
'epochs': 2,
'batch_maxnum': 100,
#...
'test_ratio' : 0.25,
'test_metric': MetricTypes.F1_SCORE,
'test_on_global_updates': True,
'test_on_local_updates': True,
'test_batch_size': 0,
'test_metric_args': {'average': 'macro'}
}
exp = Experiment(# ....
training_args=training_args)
Setting Validation Arguments using Setters of the Experiment Class
Each validation argument has its own setter method in the experiment class where federated training is managed. Therefore, validation arguments can be set, modified, or reset using the setters. To enable setters for validation arguments, the experiment should be created in advance.
from fedbiomed.common.metrics import MetricTypes
from fedbiomed.researcher.federated_workflows import Experiment
training_args = {
'optimizer_args': {
'lr': 1e-3,
},
'epochs': 2,
'batch_maxnum': 100,
'test_batch_size': 0
}
exp = Experiment(training_args=training_args)
exp.set_test_ratio(0.25)
exp.set_test_on_local_updates(True)
exp.set_test_on_global_updates(True)
exp.set_test_metric(MetricTypes.F1_SCORE) # or exp.set_test_metric('F1_SCORE')
exp.set_test_metric_args({'average': 'macro'})
Setters allow updating validation arguments from one round to others.
exp.run(rounds=2, increase=True)
exp.set_test_ratio(0.35)
exp.set_set_test_metric(MetricTypes.ACCURACY)
exp.run(rounds=2, increase=True)
Using Validation facility with heavy datasets
In some specific cases, you may have some very huge datain your datasets, such as 3D images, or you want a huge validation set, that you can load on Nodes only through batches, in order to avoid Nodes to crash due to lack of Memory. Fed-BioMed provides an option to compute validation metrics through batches, in the TrainingArgument: test_batch_size.
Validation will be computed over batches instead of computing metric using the whole dataset (which is the default behaviour) if test_batch_size is set. For now, Fed-BioMed does not provide a way to reconcile all these metrics under one metric (such as computing the mean of Accuracy for instance). If you want to do so, please consider defining your own Validation step by writing your own testing_step method in the TrainingPlan (see below for further details).
training_args = {
'optimizer_args': {
'lr': 1e-3,
},
'epochs': 2,
'test_ratio': .4,
'test_batch_size': 64 # validation metrics will be done by considering batches of size 64 each
}
exp = Experiment(training_args=training_args)
Define Custom Validation Step
Fed-BioMed training plans allow defining custom validation steps for model evaluation on the node side. The name of the method that should be defined in the training plan is testing_step. It should take two input arguments as data/inputs and target/actual that are provided on the node side. The validation step can calculate and return multiple testing metrics as long as the return value of the method is supported. The method should return:
- Single
floatorintvalue that represents a single validation result. The name of the metric will be displayed asCustom.
def testing_step(self, data, target):
# Validation actions ...
value = 1.001
return value
- List of multiple validation results. Metrics names will be displayed as
Custom_1,Custom_2,Custom_n.
def testing_step(self, data, target):
# Validation actions ...
value_1 = 1.001
value_2 = 1.002
return [value_1, value_2]
- Dictionary of multiple metric results as
intorfloat. Metrics names will be displayed as the keys of dictionary.
def testing_step(self, data, target):
# Validation actions ...
result = {'metric-1' : 0.01, 'metric-2': 0.02}
return result
Info
testing_step has a higher priority than default test metrics. It means that if both testing_step in training plan and test_metricargument in the validation arguments are defined, node will only execute the methodtesting_step`
The modules, functions, and methods that are going to be used in the validation method should be added as dependencies in the training plan (see PyTorch and Sklearn). Please also make sure that the modules whose functions will be used in the validation step do exist in the Fed-BioMed node environment.
PyTorch
The validation method in PyTorch-based training plans takes two arguments respectively for input (X) and target (y). These arguments are instances of torch.Tensor. The validation mode of PyTorch will be already activated on the node side before running testing_step with self.eval(). Therefore, there is no need to configure it again in the validation step method.
The following code snippet shows an example testing_step that calculates negative log-likelihood, cross-entropy and accuracy.
import torch
from fedbiomed.common.training_plans import TorchTrainingPlan
class MyTrainingPlan(TorchTrainingPlan):
# Other necessary methods e.g. `def init_model`
# .......
def testing_step(self, data, target):
pred = self.model().forward(data)
nll = torch.nn.functional.nll_loss(pred, target) # negative log likelihood loss
ce = torch.nn.functional.cross_entropy(pred,target) # cross entropy
_, predicted = torch.max(pred.data,1)
acc = torch.sum(predicted==target)
accuracy = acc/len(target) # accuracy
return { 'NLL': nll, 'CE': ce, 'ACCURACY': accuracy}
Info
Datasets for validation (data and target) are not a batch iterator. They contain all the samples in one block. However, it is possible to define batch iterator in the validation method as long as the method returns a single value for each metric that is calculated.
SkLearn
The validation method in scikit-learn based training plans also takes two arguments respectively for input/data (X) and target (y). These arguments are instances of np.ndarray.
The following code snippet shows an example testing_step that calculates hinge loss and accuracy.
from fedbiomed.common.training_plans import FedPerceptron
from sklearn.metrics import hinge_loss
import numpy as np
class SkLearnClassifierTrainingPlan(FedPerceptron):
def init_dependencies(self):
return ['import torch',
"from sklearn.linear_model import Perceptron",
"from torchvision import datasets, transforms",
"from torch.utils.data import DataLoader",
"from sklearn.metrics import hinge_loss"]
def compute_accuracy_for_specific_digit(self, data, target, digit: int):
idx_data_equal_to_digit = target == digit
predicted = self.model().predict(data[idx_data_equal_to_digit])
well_predicted_label = np.sum(predicted == digit) / np.sum(idx_data_equal_to_digit)
return well_predicted_label
def testing_step(self, data, target):
# hinge loss
distance_from_hyperplan = self.model().decision_function(data)
loss = hinge_loss(target, distance_from_hyperplan)
# get the accuracy only on images representing digit 1
well_predicted_label_1 = self.compute_accuracy_for_specific_digit(data, target, 1)
# Returning results as dict
return {'Hinge Loss': loss, 'Well Predicted Label 1' : well_predicted_label_1}
Conclusion
The validation part in FL plays an important role in evaluating model performance that is trained on different nodes with different datasets. Applying a validation on the node side for each training round allows comparing the impacts of particular nodes on the trained model. Understanding and comparing different impacts will be clearer thanks to two types of validations: validation on aggregated parameters and validation on locally trained parameters.


